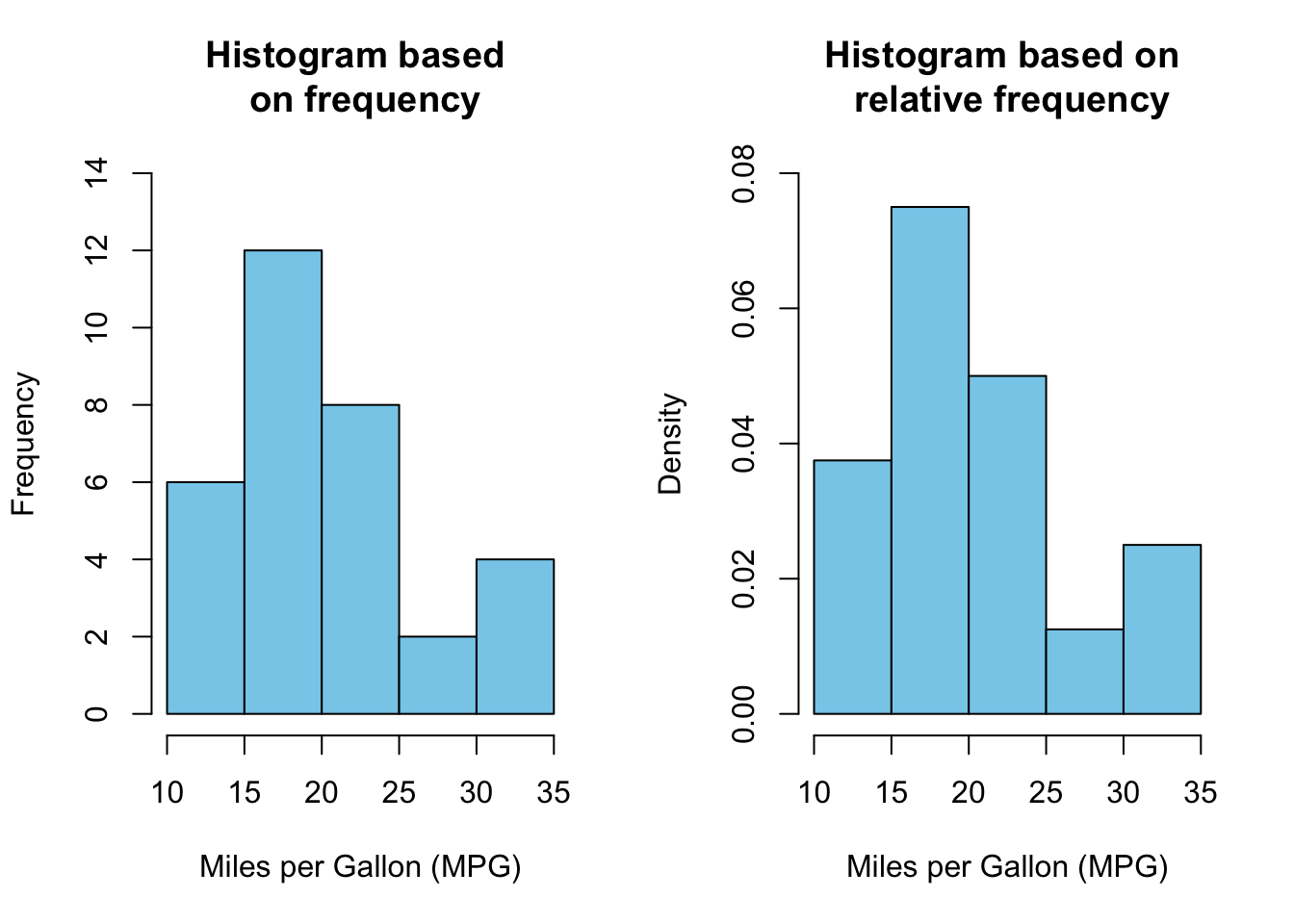
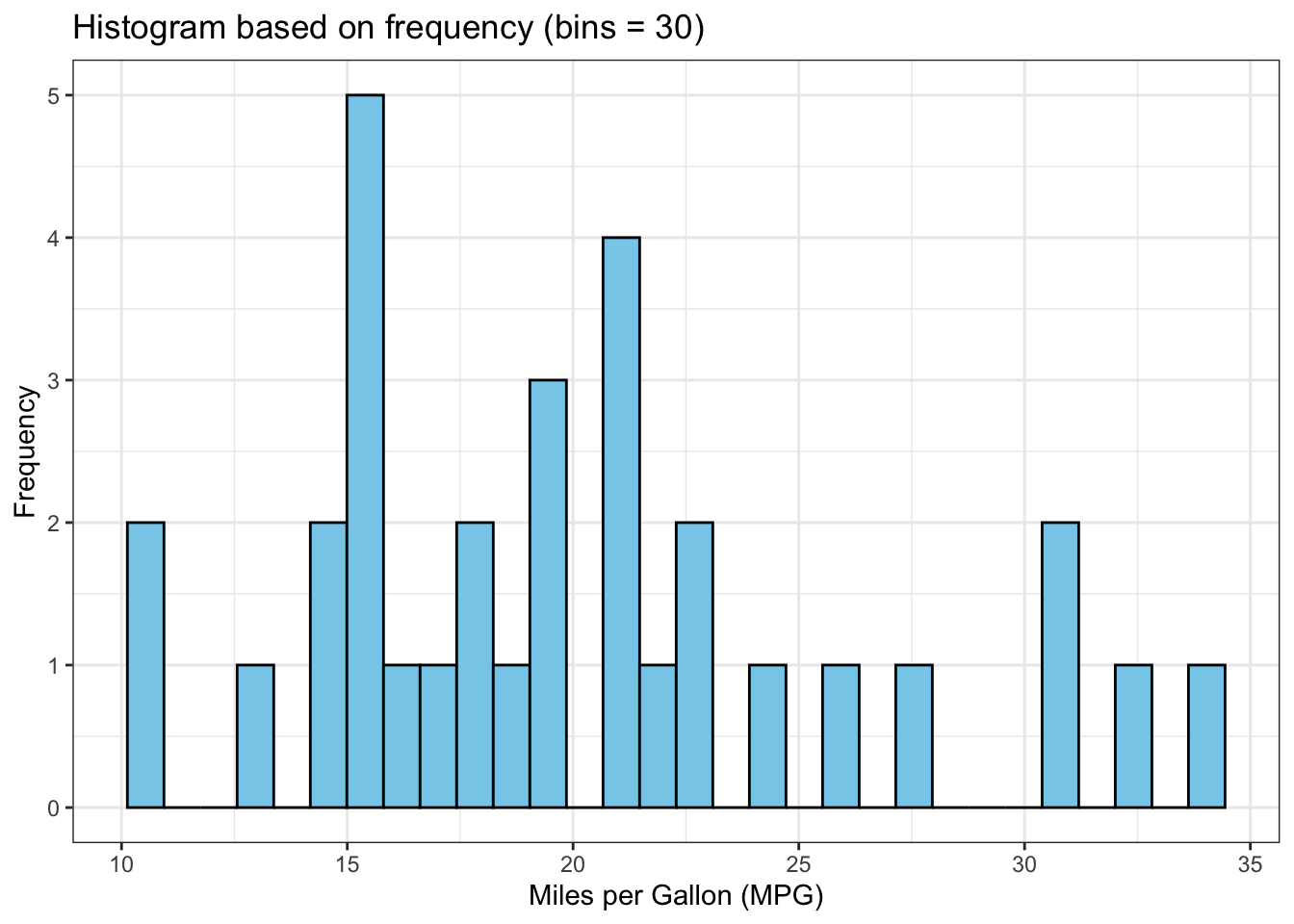
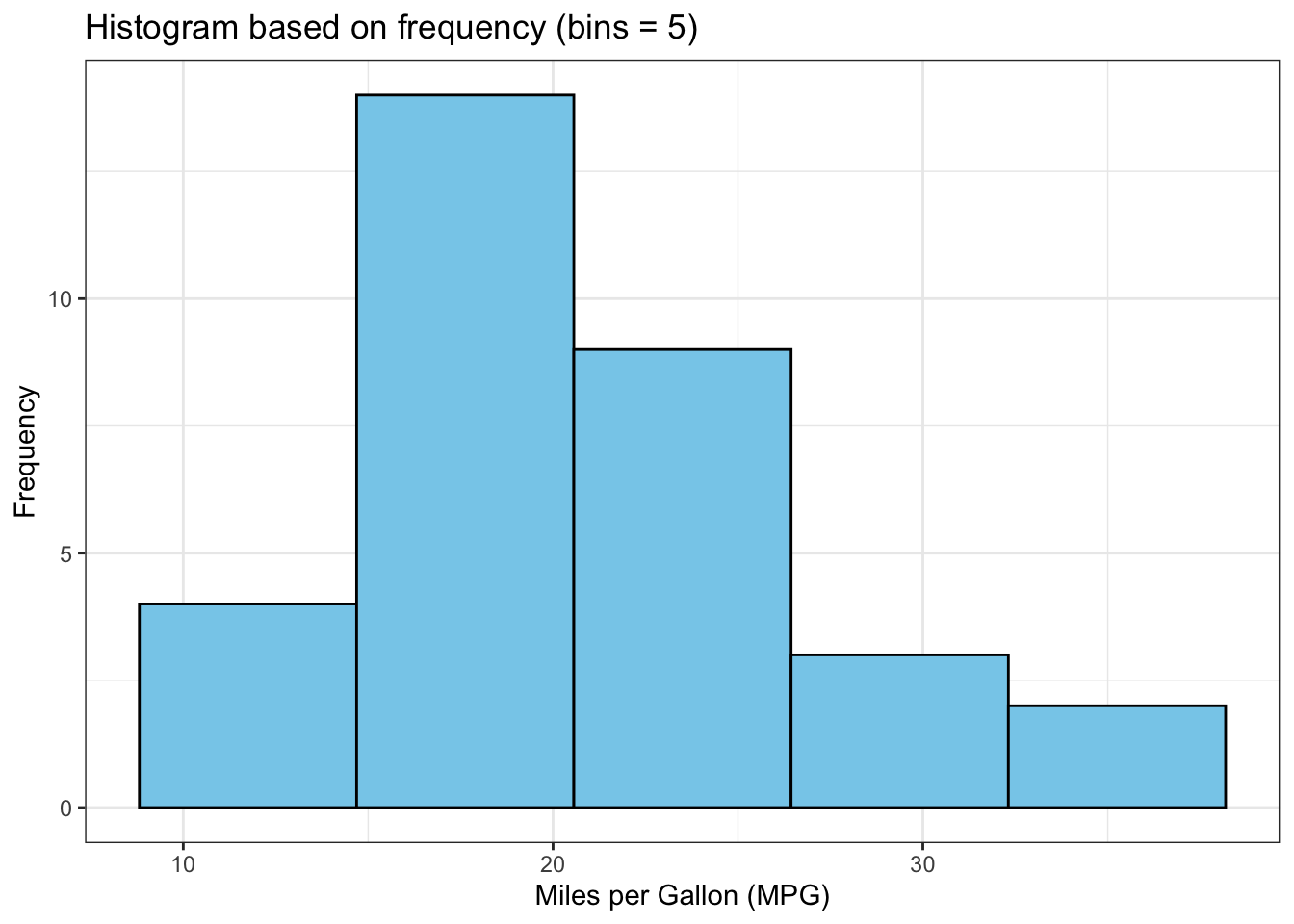
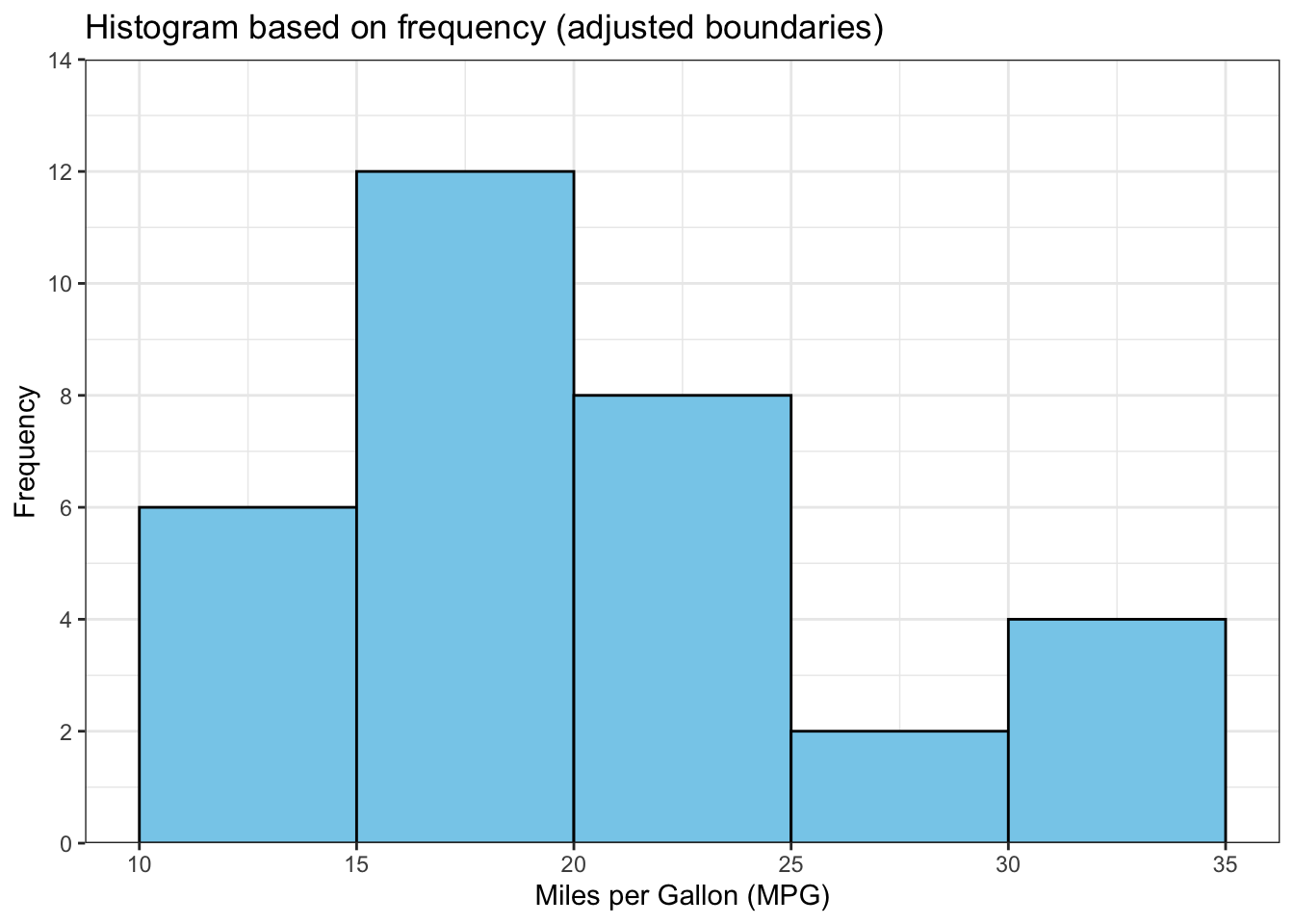
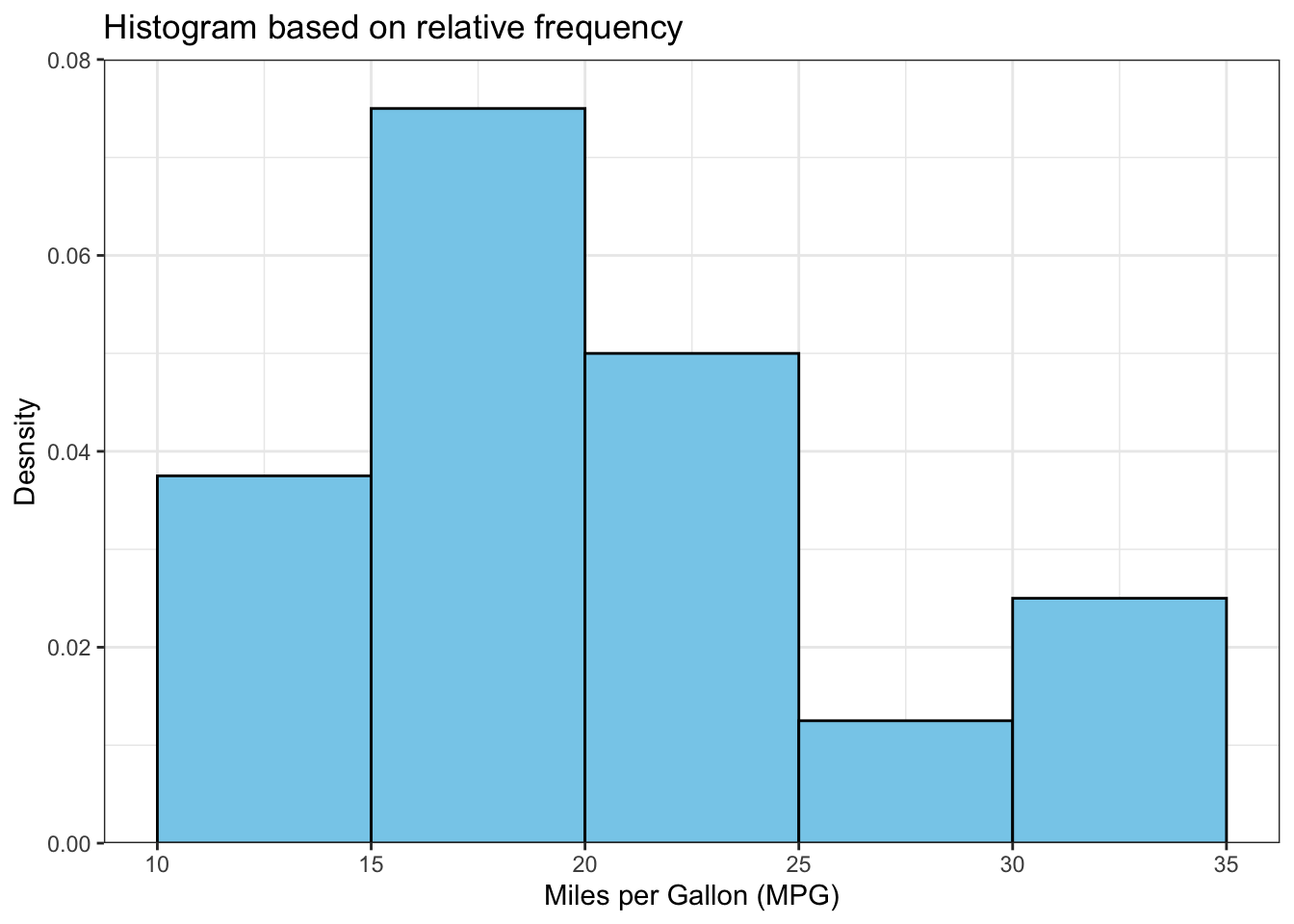
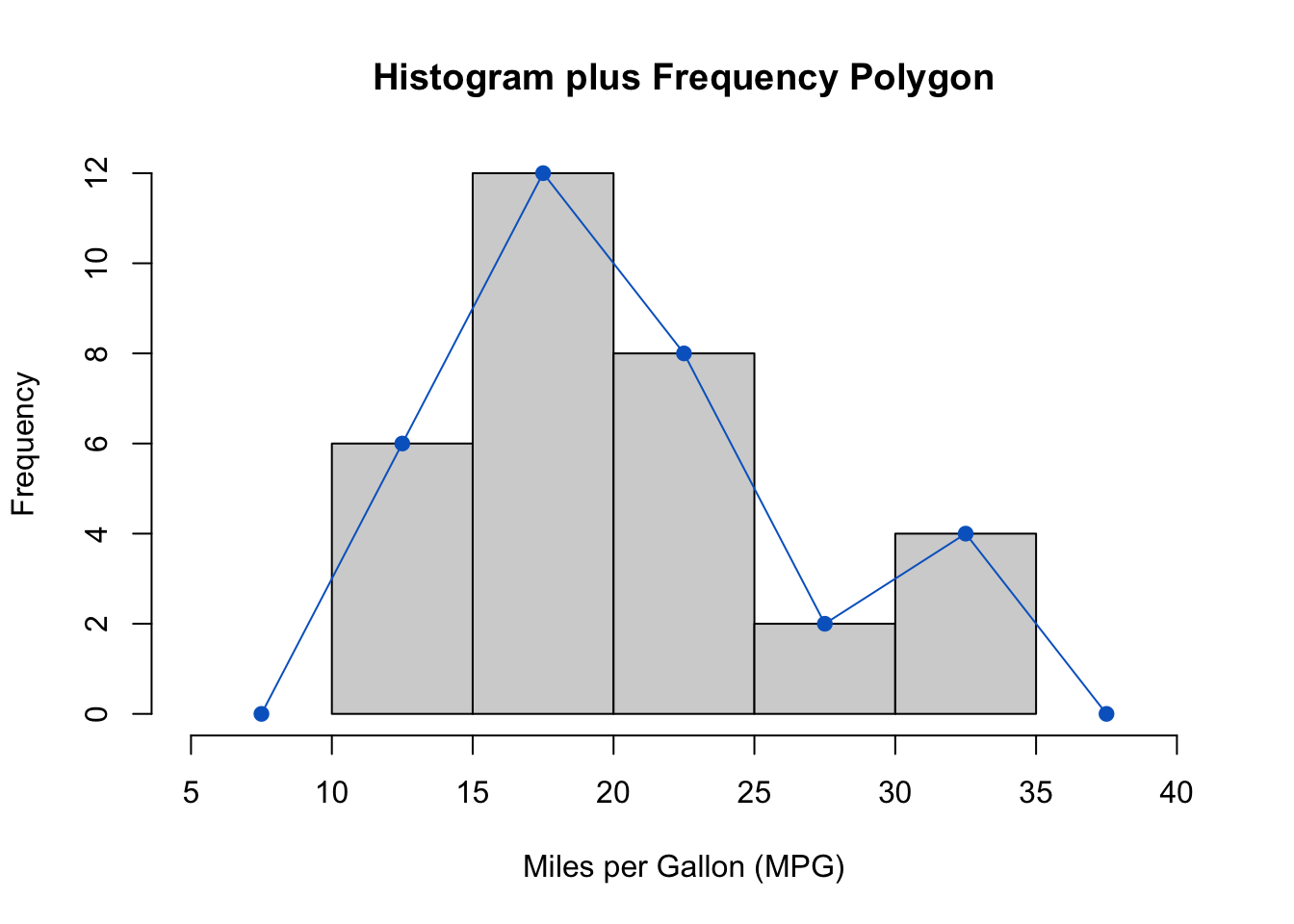
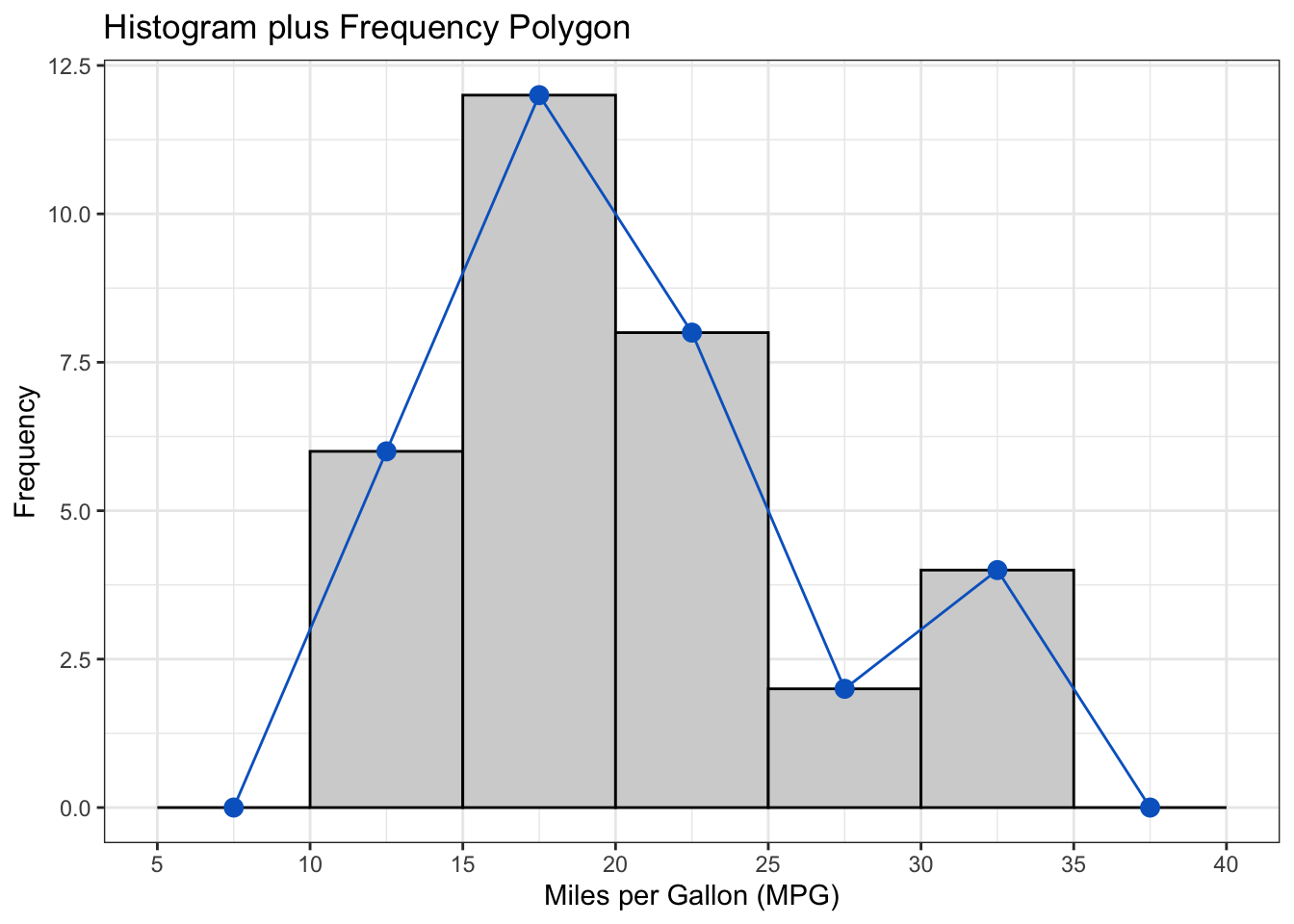
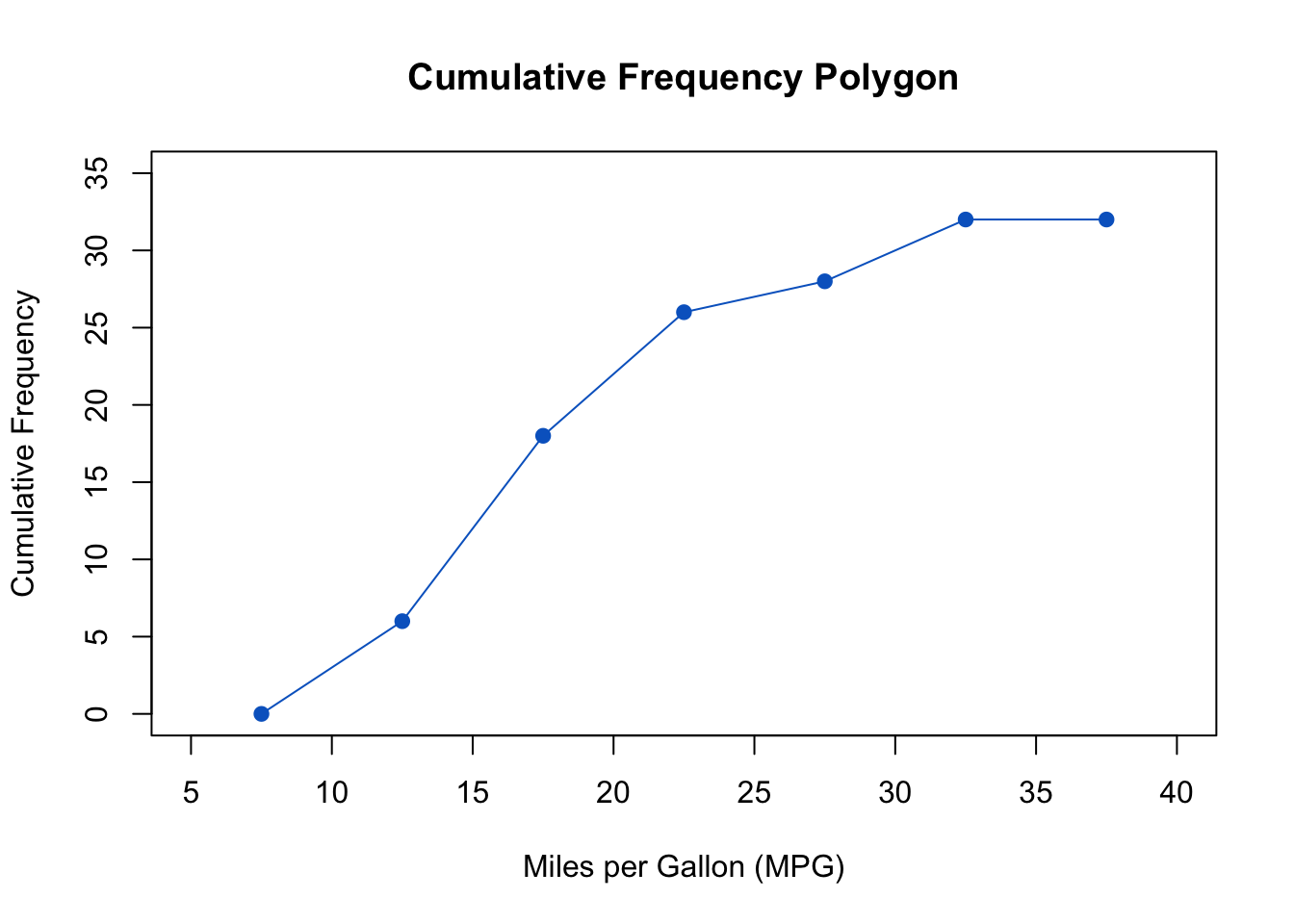
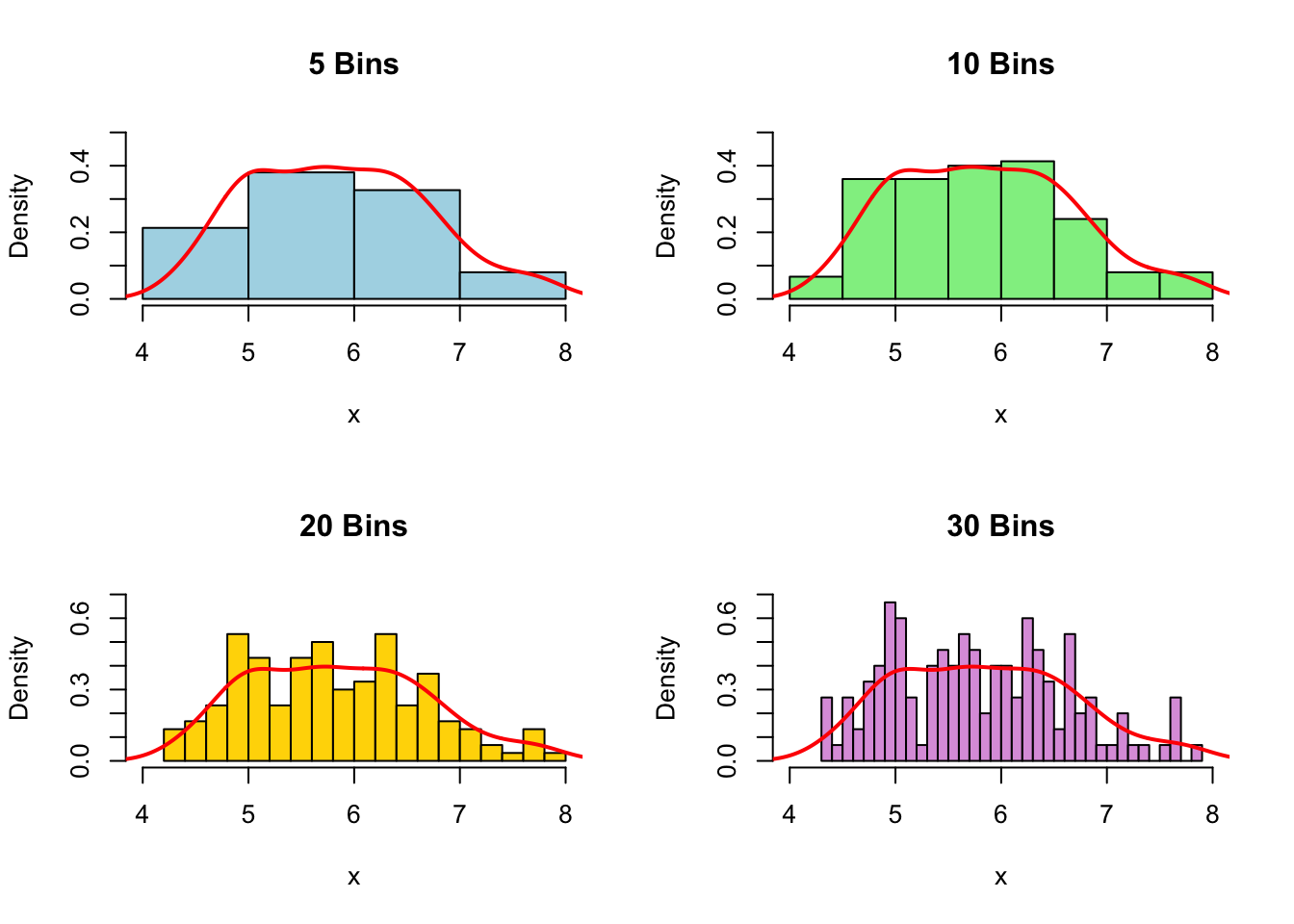
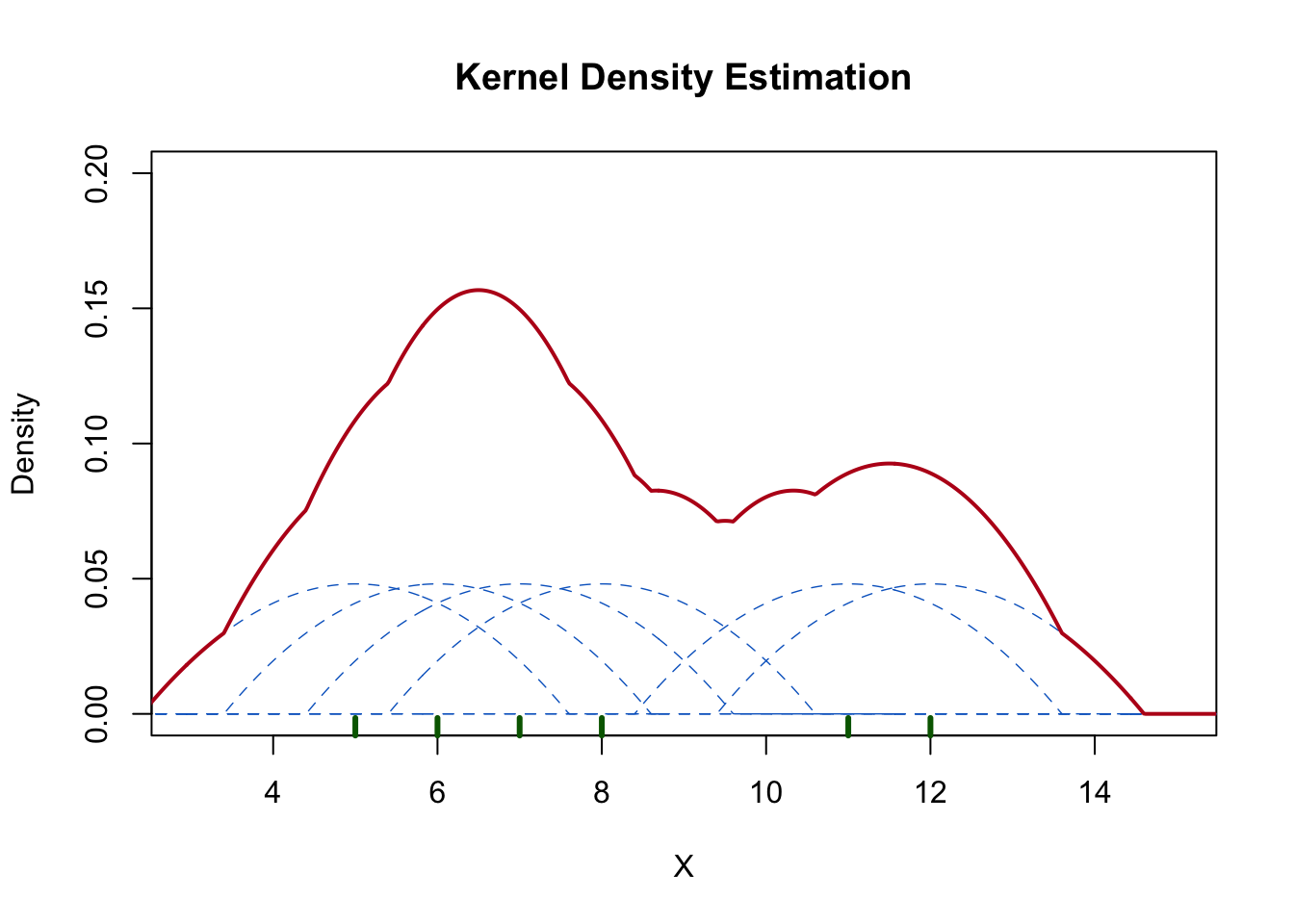
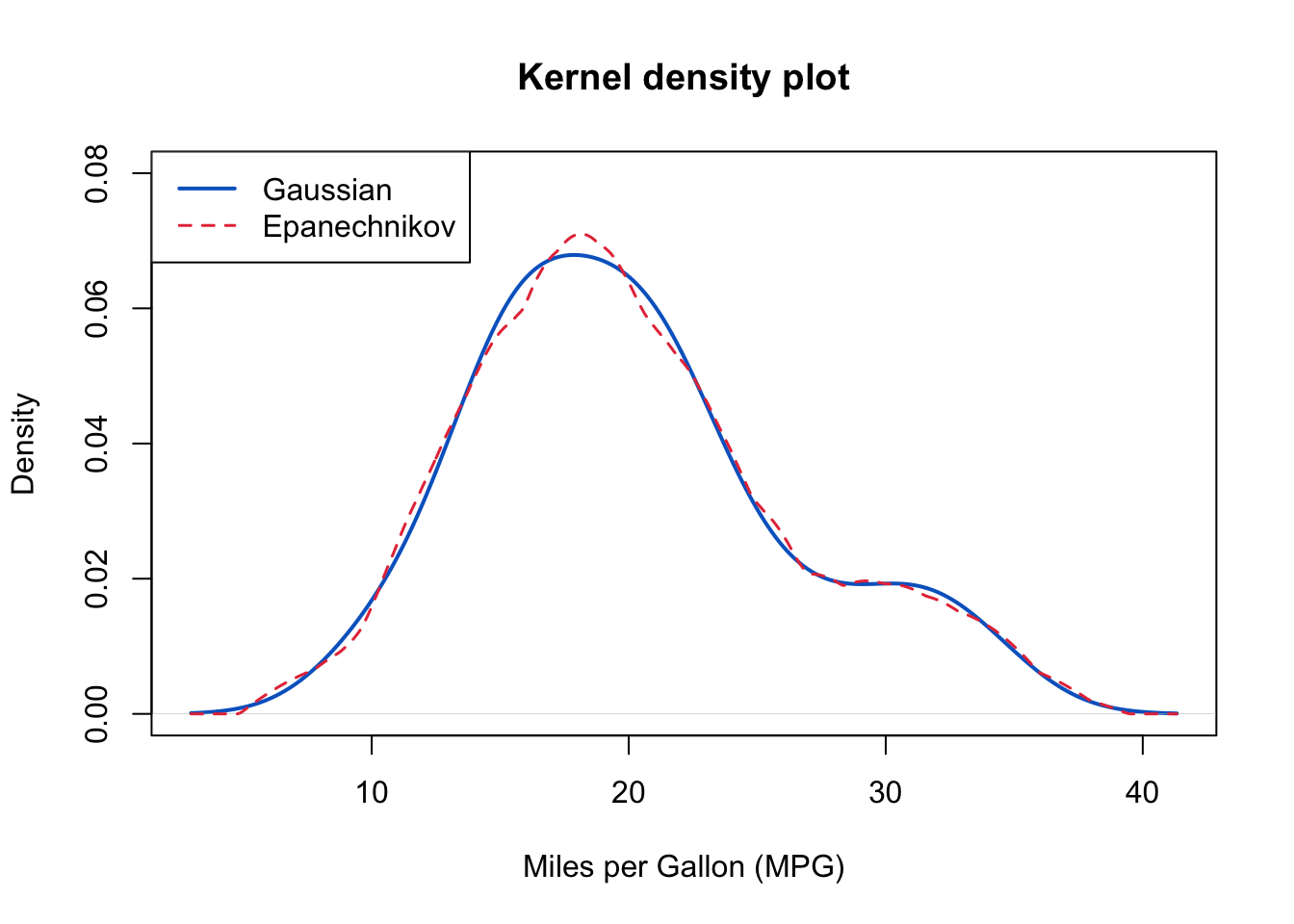
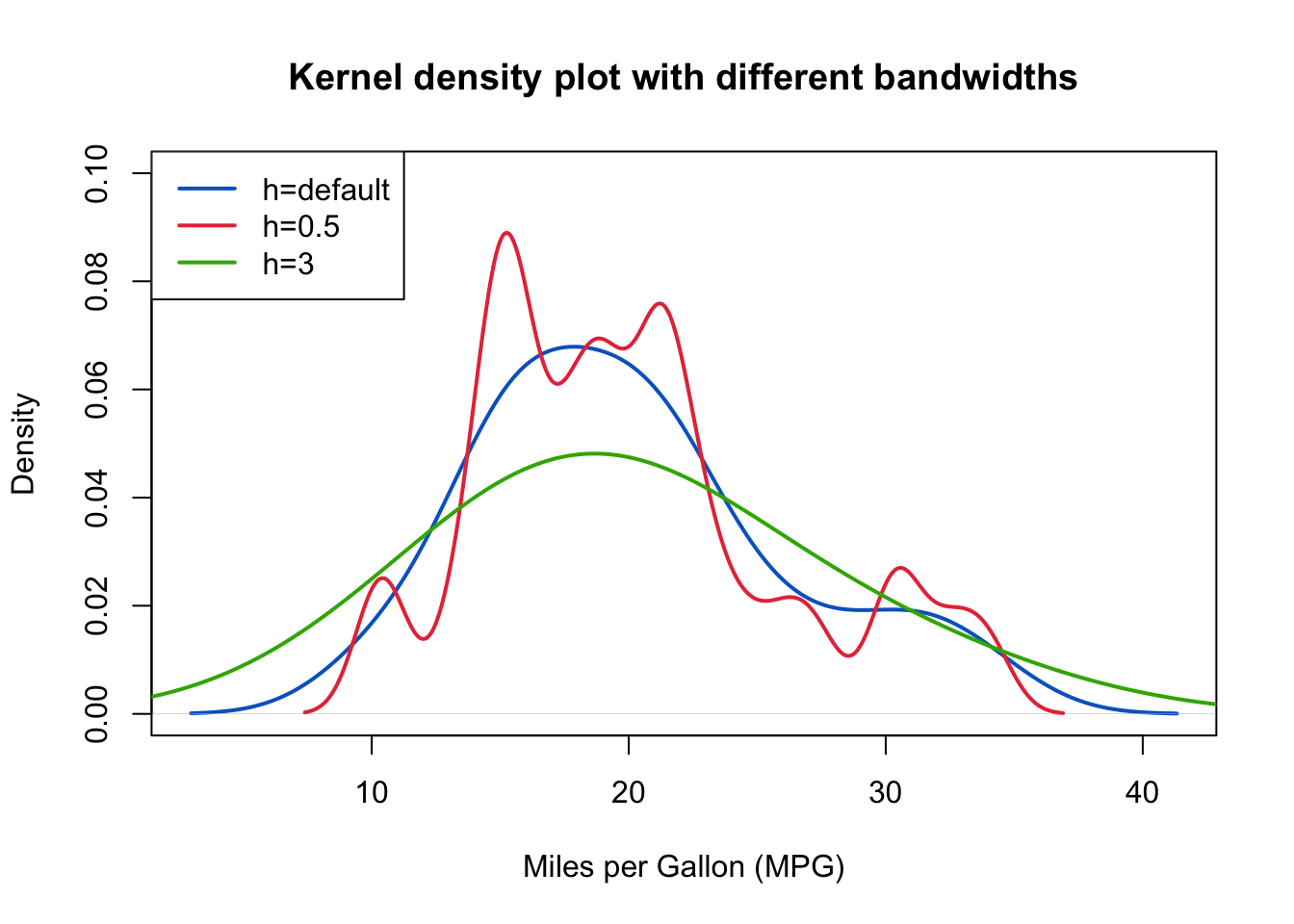
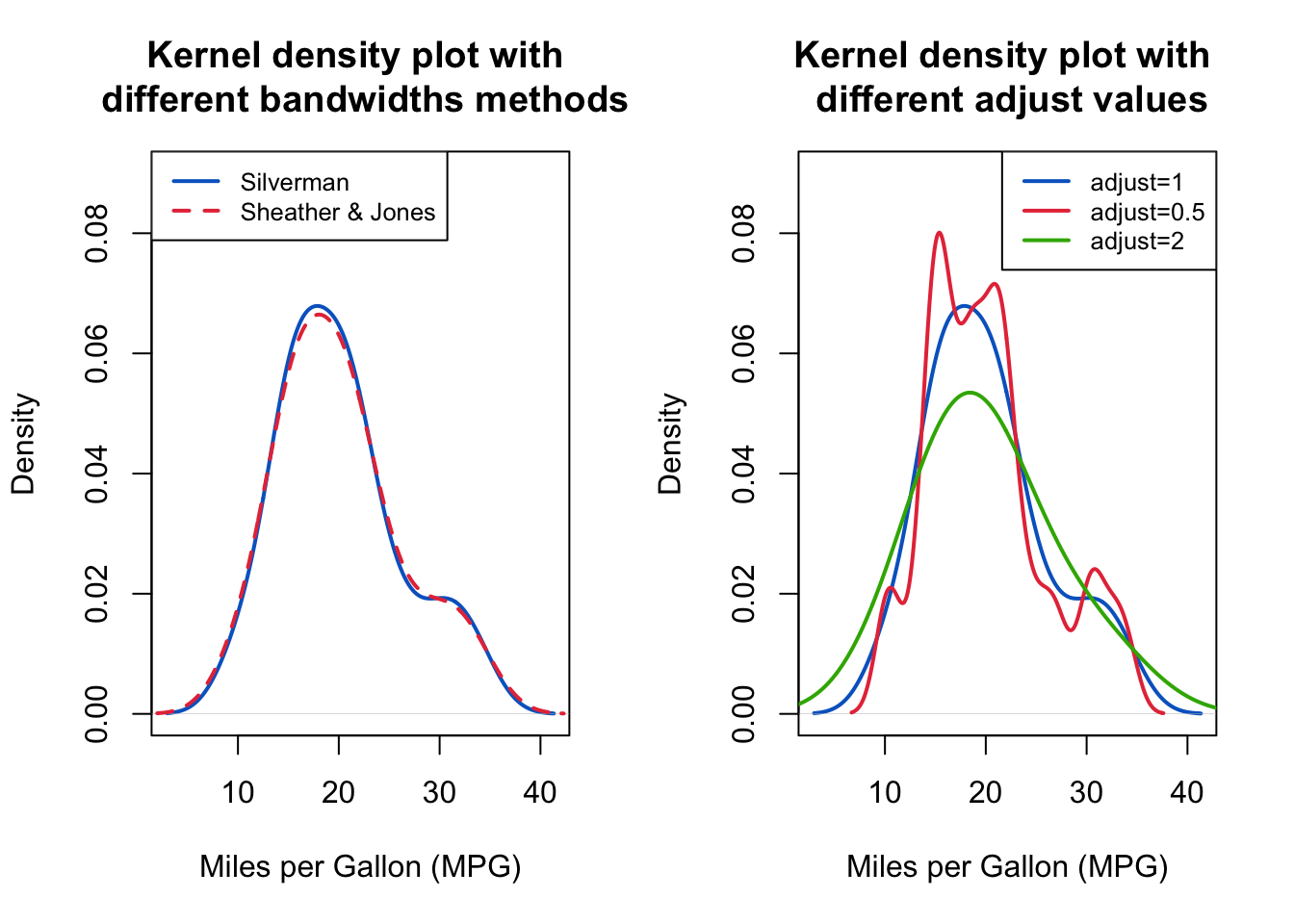
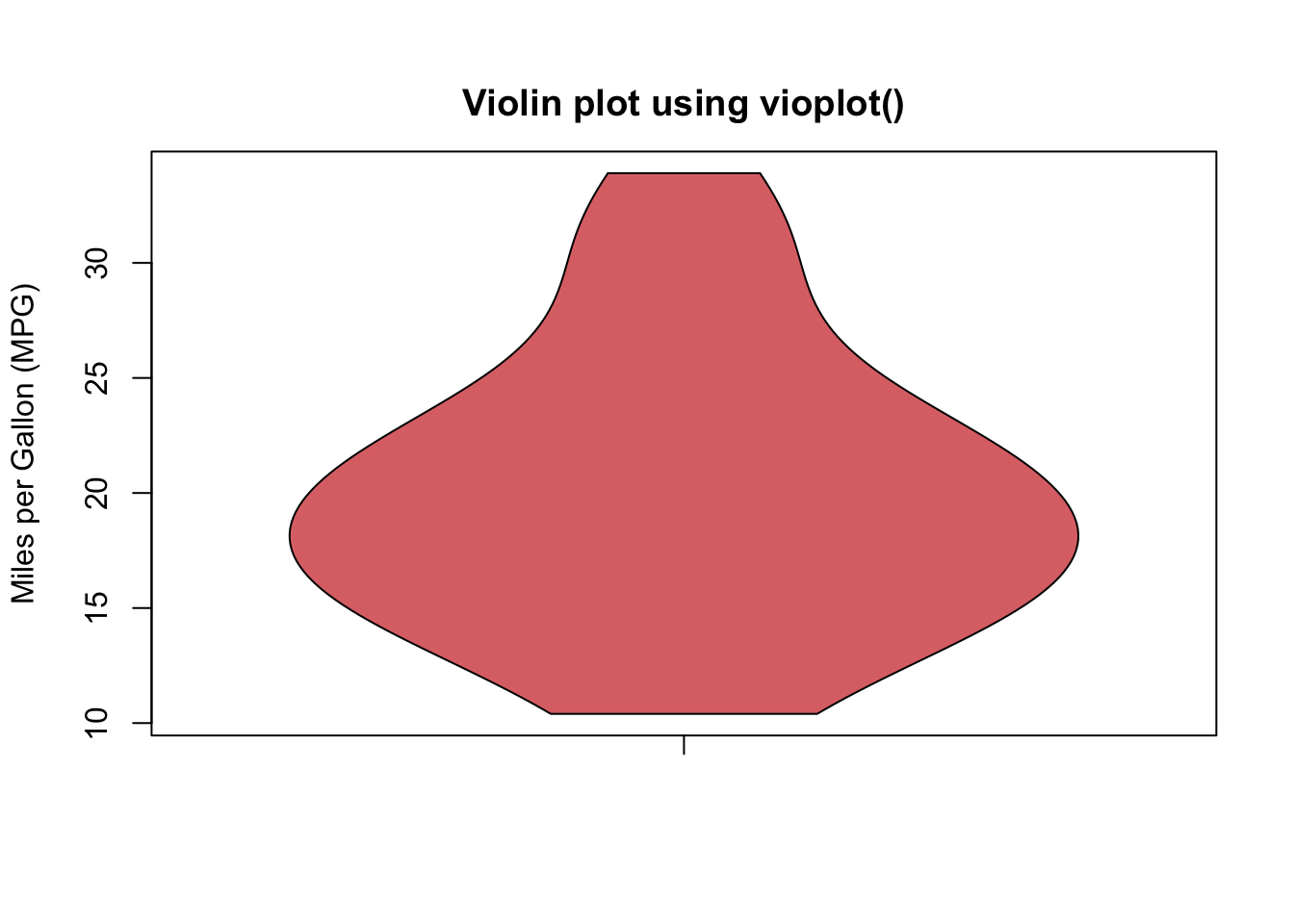
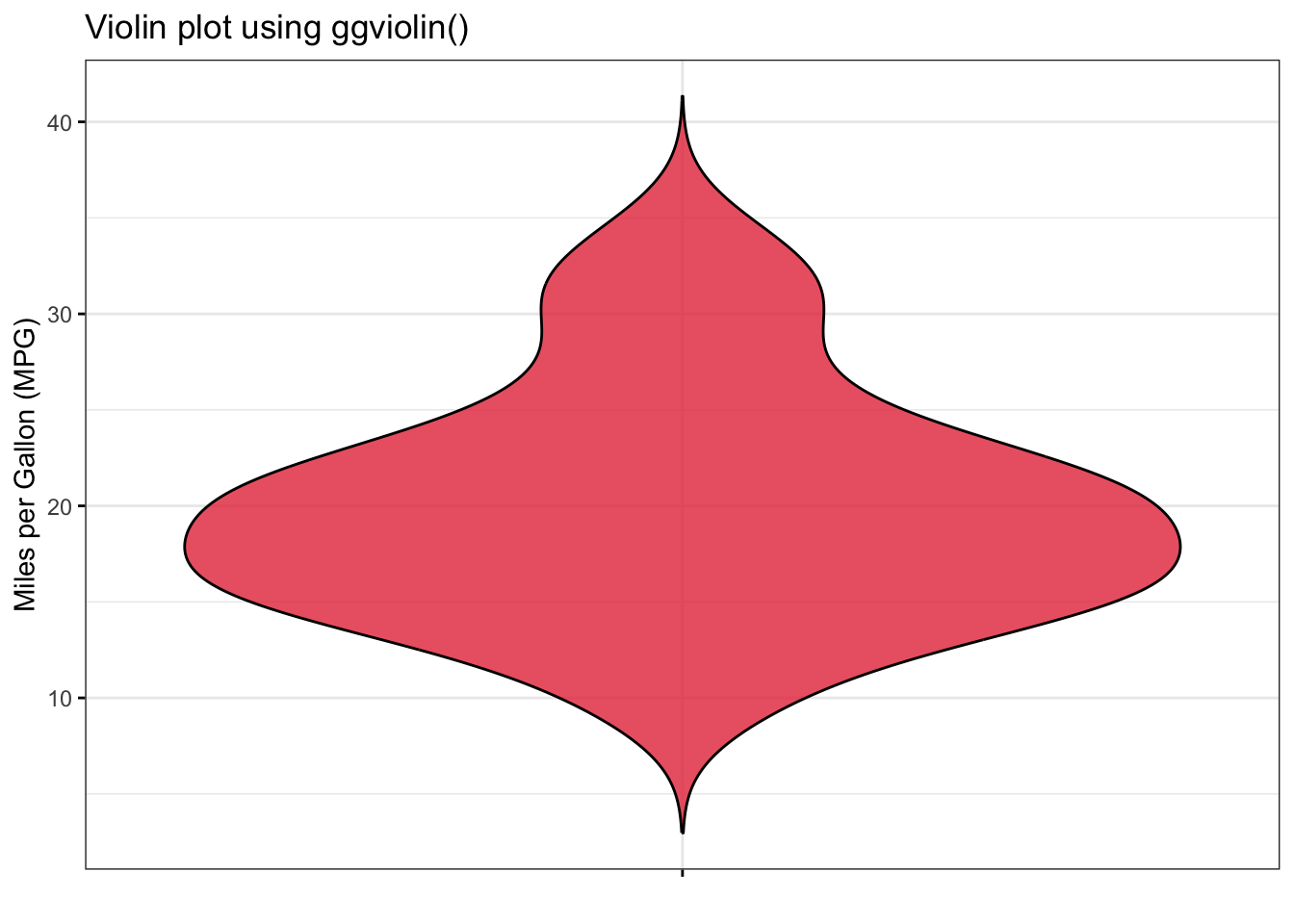
The above output can be interpreted as follows:
This header indicates that stem should be interpreted as tens (or hundreds, etc), whereas leaf should be interpreted as ones not as tenth decimal place (i.e., interpret \(1\ |\ 2\) as \(12\) not \(1.2\)). This can be controlled using the argument unit, which accepts numbers as power of 10 (e.g., 2, 1, 0.1, 0.01, etc).
This shows that the leaf unit is 1 (i.e., the leaf represents the ones place).
This shows the ntotal number of observations.
This column shows the stem values (i.e., the tens place); Each stem is separated into two parts (rows), the first contains leaves from 0 to 4, and the second contains leaves from 5 to 9.
This shows the leaves (i.e., the ones place) corresponding to the stem values.
This column is referred to as the depths, which shows the accumulated frequencies from the top and bottom of the plot until reaching the row that contains the middle value (i.e., the median). So, the last \((5^{\text{th}})\) row has depth of \(4\) because there are \(4\) observations (leaves) with stem value \(3\). The second row from the bottom \((4^{\text{th}})\) has a depth of \(6\) because there are \(4\) leaves in the \(5^{\text{th}}\) row and \(2\) leaves in the \(4^{\text{th}}\) row.
This shows the row containing the middle value (i.e., the median). The frequency of leaves in this row is placed in parentheses to indicate that the median is in this row. For example, the median of the mpg variable is \(19.2\), which is in the \(2^{\text{nd}}\) row that contains \(13\) leaves.
In the above example, the decimals have been omitted from the leaves because the unit argument is set to 1 (i.e., the leaves represent the ones place).
Sometimes, a plus \((+)\) is added at the end of one or more rows to indicate that the leaves exceed the row capacity, and thus, the leaves are not shown in the plot.